Tags
This is a joint post with my long-time friend and colleague Michelle Wille, a PhD candidate at Linnaeus University in Sweden
A recent blog post hypothesizing that the now-extinct Labrador Duck was actually not extinct, but a hybrid between two other ducks (Common Eider and Steller’s Eider) got a lot of play in some circles this week. I retweeted it, and sent it around to some waterfowl folks I know. But something just didn’t sit right.
The author, Eugene McCarthy (no, not that Eugene McCarthy), was fairly engaged on Twitter (edit: until he deleted his replies). When I pointed out that someone had done some sequencing of Labrador Duck mitochondrial DNA, though, his response was a little… odd.
The tweets have now been deleted (as were any of McCarthy’s replies/tweets to me), so this is recollection. McCarthy said that, rather than engage with Glen Chilton (arguably the leading expert on Labrador Ducks, and the author of the paper using Labrador Duck DNA), he would correspond with Bob Zink since “(Chilton) did his grad work in the department, so Bob has access to the data”. My end of the conversation is here.
Bob Zink is a well-respected ornithologist at the University of Minnesota, but that isn’t where Glen Chilton went to grad school. I became a little more skeptical of Dr. McCarthy’s claim after stumbling across this commentary on one of his earlier works, wherein he claims that humans are the result of a hybrid between pigs and chimpanzees. According to McCarthy’s website:
“During my years at the genetics department, I became increasingly dissatisfied with the standard explanation of evolution. The more I read about fossils, the more convinced I became that Darwin’s account of the evolutionary process was fundamentally flawed. Moreover, in my study of hybrids I became aware that an alternative way of thinking about evolution, what I now call “stabilization theory,” could do a better job of explaining the available data.”
McCarthy also wrote the book “Handbook of Avian Hybrids of the World” (Oxford University Press, 2006), so it would seem that he has hybrids on his mind.
So back to the question at hand – what evidence is there for a Common x Steller’s Eider hybrid being the Labrador Duck? Well, not much. Let’s start from the basics.
Eiders (and the Labrador Duck) are a group of birds known colloquially as seaducks, and are in the tribe Mergini. There are four species of eider:
- Common Eider, Somateria mollissima
- King Eider, Somateria spectabilis
- Spectacled Eider, Somateria fischeri
- Steller’s Eider, Polysticta stelleri
The most obvious thing to note is that Steller’s Eiders are in a different genus, but as Kevin Winker pointed out in his monograph on avian subspecies, “all species are hypotheses”. So let’s begin.
Morphology
McCarthy’s evidence comes from morphology and appearance. He claims that: “Indeed, in a phylogeny based on 120 separate morphological characters (Livezey 1986 – (PDF)) Camptorhynchus grouped closely with Polysticta and Somateria”. What he omits is the fact that nestled right between the two eiders, and the Labrador Duck is Histrionicus histrionicus, the Harlequin Duck, which is a separate species.
Interestingly, he seems to dismiss the actual number of specimens, writing that “(Flannery and Schouten (2001 “A gap in Nature”, Atlantic Monthly Press, New York) goes on to say that only 31 museum specimens are known (Chilton and Sorenson, 2007, state that there are 55).” In fact, Chilton visited every specimen for his book “Curse of the Labrador Duck” (Simon and Shuster, 2009). And in 1963, 54 specimens were known and their locations and histories described by Paul Hahn (“Where is that vanished bird? An index to the known specimens of the extinct and near extinct North American species”, Royal Ontario Museum and University of Toronto Press, 1963). The only one missing between Hahn and Chilton’s lists was one in a private collection on Qatar.
Nevertheless, there are two reports of Common x Steller’s Eider hybrids: one in Norway in 1995 (Forsman 1995), and one in Germany two years earlier (but first described in 1995). Here’s the description from Forsman’s sighting on 7 April 1995 in Vadsö, Norway:
“Head and neck: The head and neck were mostly white, with a dark green spot in front of the eye, which joined over the crown, and with faintly green sides to the head and neck, with the green framing and demarcating obvious white spectacles around the eyes. The bill was pale gray (paler than Steller’s Eider, and more like the colour of Common Eider).”
“Underparts: the breast was deeper orange than in Common and King Eiders, and showed a faint brown wash, like Steller’s. The colour of the breast merged into the black lower breast and belly without a clear demarcation. The ‘aft’ was all dark and the underwings pale grey”
In short, not like a Labrador Duck. (update: scroll down for the medium-quality photo)
The second sighting was in Nidersachsen, Germany, on 17 November 1993, and though photos were taken, they aren’t included in the summary (Anon. 1995). There are, though, two drawings:

The account of a Steller’s x Common Eider hybrid in Germany. From Anon. 1995. Seltene Vogelarten in Deutschland 1993. Limicola 9: 77-110
What’s obvious is the dark black on the side and back of the head, which is far more extensive than anything observed on any of the male Labrador Duck specimens (or any of the drawings or paintings).
But these are sight/photographic records, and could represent aberrant plumages, or genuine hybrids – we’ll never really know. Kenn Kaufman also succinctly covers the problems with the morphological argument. What about female Labrador Ducks? McCarthy posits that they are the hybrids of another species pair.
But I hear you saying “what about genetics?” I’m glad you asked.
Genetics
In 2007, Glen Chilton and Michael Sorenson published a paper wherein they tested whether certain eggs belonged to Labrador Ducks (they didn’t). What that paper also included, though, was the GenBank accession numbers to two Labrador Duck sequences – the 12S ribosomal subunit, and NADH2 subunit, both mitochondrial genes from a male Labrador Duck at the University of Michigan Museum of Zoology (UMMZ 152253). Well, parts of them.

Figure 1: 12S sequence from Labrador Duck compared to full length 12S Sequence (Extracted from full MtDNA of Mallard)

Figure 2: NADH2 (=ND2) sequence from Labrador Duck compared to full length NADH Sequence (Extracted from full MtDNA of Mallard)
These are the only Labrador Duck sequences in GenBank. Now, in order to do a comparison, we need these genes to be sequences in our two putative parent species – Common and Steller’s Eider. What do we have?
| Species | NADH2 | 12S |
| Polysticta stelleri | no | no |
| Somateria mollissima | yes | no |
| Somateria fischeri | no | yes |
| Somateria spectabilis | yes | no |
| Histrionicus histrionicus | no | yes |
| Melanitta nigra | yes | no |
| Melanitta americana | no | no |
| Melanitta fusca | no | no |
| Melanitta deglandi | no | no |
| Melanitta perspicillata | yes | no |
| Clangula hyemalis | yes | no |
| Bucephala clangula | yes | yes |
| Bucephala islandica | yes | no |
| Bucephala albeola | yes | no |
| Mergellus albellus | yes | yes |
| Lophodytes cucullatus | yes | no |
| Mergus octosetaceus | no | no |
| Mergus australis | no | no |
| Mergus serrator | yes | yes |
| Mergus merganser | yes | yes |
| Mergus squamatus | yes | yes |
This also includes some of the other seaducks to give you an idea of what’s out there. It’s not great, but let’s push on working with what we have. There are a bunch of Steller’s Eider sequences in GenBank, but not for 12S or NADH2 (CO1: 3, cytb: 17, haemoglobin alpha: 1, haemoglobin beta: 1). The cytochrome b (cytb) sequences, while there are 17 of them, are not of good quality with many sites including N’s (i.e., there could be an A, G, T, or C at that position), which are unusable in our type of analysis.

Figure 3: Screenshot of a section of the P. stelleri CytB sequences in an alignment, with many N’s highlighted in grey. There is a high prevalence of unusable sites both at the start of the alignment and at the end.
But the CO1 (cytochrome oxidase 1) sequences are of better quality (no N’s!) and we have better coverage of the gene (617bp of the 1500bp gene).
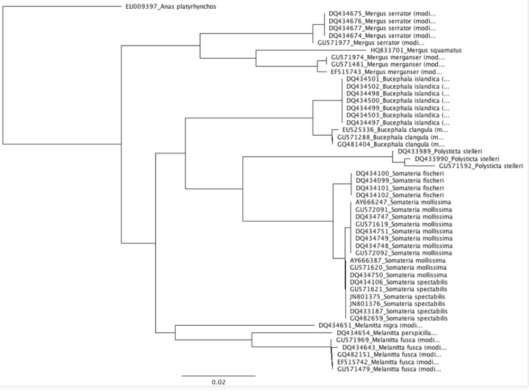
Figure 4: Garli Tree (maximum likelihood) of a 617bp region of the COI gene of the Mergini Tribe with Mallard as an outgroup.
While this is a quick and dirty tree, it distinguishes the different seaduck species into discrete clades. In terms of the eiders, Steller’s Eiders (Polysticta stelleri) are an outgroup to the Somateria eiders, and S. fischeri is a sister species to S. mollissima and S. spectabilis. To generate robust phylogenies we’d want to include a number of (preferably full length) genes. Case in point, S. mollissima and S. spectabilis are not discrete clades in our figure, because there is only one single nucleotide polymorphism (SNP) separating the two species in the region used for the analysis. The reason for overlap is that not all S. molissima have this SNP. However, there is enough data in this analysis to distinguish Steller’s and Common Eiders as species (discrete clades in the tree and 93% pairwise identity in sequences of the two species).
OK – so to recap, both Common and Steller’s Eiders are valid species that are genetically quite different. Now, where do Labrador Ducks fit in? Let’s got back to the Labrador Duck 12S and NADH2 genetic data.
The only regions we can keep in the analysis are those where there is sequence from all the species in question. We could remove species for which the sequences are too short, or have poor overlap, however, in terms of the NADH gene, that would require removing sequence from the Eiders, which we are the most interested in.

Figure 5: Nucleotide alignment of the NADH region where in we only have overlap of 33bp, if we retain sequence 4, and 51bp if we remove sequence 4 and retain sequence 5, which is the only representative of S. spectabilis.
While these sequences are informative enough for the analysis done by Chilton and Sorenson, they are not informative enough to build phylogenetic trees. In order to place Labrador Ducks in any phylogeny, longer sequences are required, and/or inclusion of multiple genes.
Gonzalez et al. (2009) built a pretty robust tree based on 2 full length mitochondrial genes (NADH and Cytb), concatenated (where the sequences of 2 genes are combined to produce a longer stretch of DNA) to produce and alignment over 2000bps. We have 137bp (12S) and 72bp (NADH), before trimming, and as we don’t have sequence for both regions in most species, we can’t combine them to get us up to 200bp (Table 1).
But, what we can do is use an approach similar to Chilton and Sorenson to see if the Labrador Duck sequences are highly similar to those of Eiders. So, if we take out alignments, trim them, and calculate pairwise identity, we can get an idea of how similar our sequences are to those of other Anatidae.
Table 2: Percentage identity of 12S mtDNA (122bp alignment).
| Name (GenBank Accession_Species) | % Identity |
| DQ831207_Camptorhynchus labradorius | 100.0 |
| DQ831211_Mergus serrator | 94.2 |
| DQ831212_Mergus serrator | 94.2 |
| AF173687_Anas gibberifrons | 93.4 |
| AF173481_Anas castanea | 93.4 |
| AF173712_Bucephala clangula | 93.4 |
| AF173485_Anas platyrhynchos | 93.4 |
| AY164517_Anas poecilorhyncha | 93.4 |
| AF173486_Anas superciliosa | 93.4 |
| HM063550_Cyanochen cyanopterus | 93.4 |
| U83738_Somateria fischeri | 93.4 |
| AF173708_Tadorna radjah | 93.4 |
| S76217_Cygnus melancoryphus | 93.4 |
| AF173487_Anas chlorotis peculiaris | 92.9 |
| AY164518_Anas penelope | 92.7 |
| AY164527_Anas falcata | 92.7 |
| AF173691_Anas querquedula | 92.7 |
| AF173702_Sarkidiornis melanotos | 92.7 |
| HM063541_Sarkidiornis melanotos | 92.7 |
| AY164528_Aythya fuligula | 92.7 |
| HM063556_Merganetta armata | 92.7 |
| AF173706_Tadorna variegata | 92.7 |
| AF173707_Tadorna tadorna | 92.7 |
| AF173480_Anas gracilis | 92.0 |
| AF173689_Anas strepera | 92.0 |
| AF173692_Anas hottentota | 92.0 |
| AF173711_Mergellus albellus | 92.0 |
| AY164529_Mergus merganser | 92.0 |
| HM063548_Asarcornis scutulata | 92.0 |
| HM063554_Neochen jubata | 92.0 |
| S76216_Coscoroba coscoroba | 92.0 |
| AF173703_Pteronetta hartlaubi | 91.2 |
| HM063552_Hymenolaimus malacorhynchos | 91.2 |
| AF173699_Netta rufina | 91.2 |
| AF173713_Histrionicus histrionicus | 89.8 |
| HM063555_Chloephaga melanoptera | 89.8 |
| HM063546_Callonetta leucophrys | 89.1 |
| FJ379295_Cygnus atratus | 89.1 |
| HM063545_Chenonetta jubata | 88.3 |
| AY747699_Oxyura vittata | 87.6 |
| HQ833701_Mergus squamatus* | 78.8 |
- Multiple sequences from the same species have been removed to make the table a little smaller.
- Multiple sequences from the same species are within 99% identity
- There are three sequences from Mergus squamatus, and in the alignment there is a strange deletion of 15bp, which is affecting the % identity analysis. Stripping these sites from the alignment the %identity becomes 88%. These 15bp were retained to maximize the number of sites in the alignment for comparison overall.
Table 3: Percentage identity of NADH2 (51bp).
| Name (GenBank Accession_Species) | % Identity |
| DQ831199_Camptorhynchus labradorius | 100.0 |
| AF059131_Anas flavirostris flavirostris | 96.1 |
| AF059132_Anas flavirostris oxyptera | 96.1 |
| AF059140_Anas melleri | 96.1 |
| AF059121_Anas bernieri | 94.1 |
| EU585701_Clangula hyemalis | 94.1 |
| DQ831203_Somateria mollissima | 92.2 |
| EU585725_Somateria spectabilis | 92.2 |
| DQ831200_Anas platyrhynchos | 92.2 |
| AF059163_Anas americana | 92.2 |
| AF059168_Anas sibilatrix | 92.2 |
| HM063562_Hymenolaimus malacorhynchos | 88.2 |
| AF515267_Melanitta nigra | 88.2 |
| AF515265_Bucephala clangula | 88.2 |
| EU585716_Mergellus albellus | 88.2 |
| DQ831206_Mergus serrator | 84.3 |
| HM063566_Merganetta armata | 84.3 |
| EU585717_Mergus merganser merganser | 84.3 |
| EU585715_Melanitta perspicillata | 84.3 |
| HQ833701_Mergus squamatus | 82.4 |
| AY747869_Oxyura maccoa | 82.4 |
| AY747866_Oxyura jamaicensis jamaicensis | 80.4 |
| AY747864_Nomonyx dominica | 78.4 |
| HM063568_Cygnus olor | 76.5 |
| FJ379295_Cygnus atratus | 74.5 |
- Multiple sequences from the same species have been removed to make the table a little smaller.
- Multiple sequences from the same species are within 99% identity
- Many species had to be excluded as the sequences did not overlap with a sufficient number of bases with the sequence from the Labrador Duck.
The tables demonstrate that the data are not overly informative as we would like. Based on phylogenies, the sequences from the Tribe Mergini should cluster near the top, or at the very least together, as they are more closely related to each other than to sequences from any other Tribe in the Anatidae. Rather, these species are scattered in the tables.
However, the sequences from the Labrador Ducks are 100% identical to themselves, and the next closest sequence is only 94% (12S) and 96% (NADH) identical. Based on duplicates included in the analysis (not shown), multiple sequences from the same species are within 99%. Thus, if the Labrador Duck were a hybrid of Steller’s and Common Eiders we would expect the sequences to be within 99% identity to either of these species in at least one of the two genes. This is not the case, in fact the most similar sequences are not those from eiders, but of mergansers (Beware! We can only use these regions to say what the Labrador Duck is not, not what it is similar to).
Labrador Ducks are not hybrids of Eiders.
Stop the presses!
As we were getting ready to publish this post, McCarthy added a lengthy explanation of the genetic data in Chilton and Sorenson 2007 as a sidebar to his original piece. McCarthy points out the lack of 12S and NADH2 sequences for Steller’s Eider (which we also address). These are mitochondrial genes, so would match the mother of any hybrid (because all mitochondria are inherited from the mother). Since the NADH2 sequence didn’t match that for Common Eider, we can rule out a female Common/male Steller’s Eider pairing. McCarthy posits that the Labrador Duck’s size (smaller than a Common Eider, larger than Steller’s) means that the Steller’s Eider is the mother. This is doubtful based on the two documented hybrids noted above, but without adequate genetic data, it remains a possibility (though a slim one).
On the issue of within-species variability, sequences that are more than ~98% different are considered separate species (compare the values in Tables 2 and 3, for example). Any within-species genetic variance is massively smaller than between-species variance. This also addresses McCarthy’s final issue with the data in Chilton and Sorenson:
“Another problem is that Chilton and Sorenson took their Labrador Duck sample from a single feather. From the standpoint of reliability, it would have been preferable to obtain sequences from other Labrador Duck specimens to confirm their sequences. That is, for PCR to be reliable, multiple sequences must (1) be generated with separate amplifications; and (2) cross checked with each other to eliminate phantom sequences resulting from contamination (which is a very common problem with that technique). Given the rarity of museum specimens, it’s understandable that they chose not to do this, but without such cross comparison, such data is next to useless.”
While he makes good points, the data are not useless. If these data were useless, the paper by Chilton and Sorenseon would never have been accepted, and we are using the same principle here as they did in that paper. However, he is correct in that amplification from low concentration, highly fragmented DNA has problems including allelic dropout, contamination, or artefacts that can compromise estimates of variability and population size (Taberlet et al. 1999, Sefc et al 2003, Buchan et al. 2005). Additionally, artificial SNP’s most often include C->T substitutions (Sefc et al 2007). These problems have very large implications for analyses done at population level, however, this is not a population level inquiry.
Additionally, a closer look at the alignment doesn’t suggest the introduction of SNP’s that only occur in the Labrador Duck sequences (ie, artefact SNPs), with an exception in the 12S where there is a unique C->T substitution (We’re happy to make the alignments available to anyone who wants them). While scanning alignments isn’t overly scientific, without seeing the original sequences this is as good as it gets.
Further, scrutiny of the methods presented in the paper by Chilton and Sorenson suggest that the amplification of sequences was completed prior to any work on the eggs to limit the possibility of contamination. The authors were very aware of the possibility of contamination and took steps to control for that. Sorenson is also something of an expert in working with ancient DNA, and has authored or coauthored many papers on error rates.
Most labs that do this type of work have negative control and are very aware of possibility of contamination. Further, methods utilized in this paper are those developed and utilized for museum specimens (Sefc et al 2003, Sorenson and Payne 2005). While the sequences have limited utility, as mentioned both in this post and in McCarthy’s response, suggesting “the data is next to useless” is hyperbolic.
Based on the very limited genetic data available, and the two known cases of Steller’s Eider x Common Eider hybrids, we propose that Labrador Ducks are not hybrids of these two species. However, we do recognize that the data available are limited, and strongly recommend sequencing (and resequencing) a number of genes from multiple feathers from multiple individuals to increase our certainty.
References
Anon. 1995. Seltene Vogelarten in Deutschland 1993. Limicola 9: 77-110.
Buchan et al. 2005. Locus effects and sources of error in noninvasive genotyping. Molecular Ecology Notes. 5:680-683
Chilton, G. 2009. The curse of the Labrador Duck: my obsessive quest to the edge of extinction. Simon & Shuster, Toronto.
Chilton, G. & M.D. Sorenson. 2007. Genetic identification ofeggs purportedly from the extinct Labrador duck. The Auk 124: 962-968.
Forsman, D. 1995. A presumed hybrid Steller’s Eider x Common Eider in Norway. Birding World 8(4): 138.
Gonzalez, J. et al. 2009. Phylogenetic relationships based on two mitochondrial genes and hybridization patterns in Anatidae. Journal of Zoology 279: 310-318
Livezey, B.C. 1986. A phylogenetic analysis of recent anseriform genera using morphological characters. Auk 103: 737-754.
McCarthy, E.M. 2006. Handbook of avian hybrids of the world. Oxford University Press, Oxford, UK.
Sefc et al. 2007. Single base errors in PCR products from avian museum specimens and their effect on estimates of historical genetic diversity. Conservation Genetics 8:879-884
Sefc et al. 2003. Microsatellite amplification from muserum feather samples: effects of fragment size and template concentration on genotyping errors. The Auk 120: 982-989
Taberlet et al. 1999. Noninvasive genetic sampling: look before you leap. Trends in Ecology and Evolution 14:323-327
Sorenson, M.D. & R.B. Payne. 2005. Molecular systematics: cuckoo phylogeny inferred from mitochondrial DNA sequences. Pp. 68-94 in R.B. Payne. Bird Families of the World: Cuckoos. Oxford University Press, Oxford, UK.
Update
27 Feb 2014 – Here’s the best photo of the 1995 Norwegian Common x Steller’s Eider hybrid.


Pingback: Friday recommended reads #19 | Small Pond Science
Excellent article to debunk the original paper, must admit most of it is beyond my understanding but you guys obviously understand these things well enough to remove doubt.
Pingback: FAQ, and answers thereto #2 « The Lab and Field